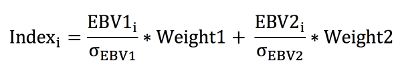The parameter file outlined below illustrates how to simulate a population undergoing selection based on an index value, which is comprised of the estimated
breeding values (ebv) for trait 1 and 2. The estimated breeding values for trait 1 and trait 2 are predicted from a bivariate pedigree (pblup) or single-step genomic BLUP
(ssgblup) model. A pblup bivariate animal model was utilized from generations 4 to 10. Starting at generation 10 all parents in the current population and selection candidates
were genotyped and for the remaining generations a ssgblup bivariate animal model was utilized. Similar to Example 6, the index for animal `i' is calculated as follows:
−−−−−−−| Running the Program Example |−−−−−−−
−| General |−
START: founder
SEED: 1500
−| Genome & Marker |−
CHR: 3
CHR_LENGTH: 150 150 150
NUM_MARK: 4000 4000 4000
QTL: 150 150 150
−| Population |−
FOUNDER_Effective_Size: Ne70
MALE_FEMALE_FOUNDER: 50 400 random 3
VARIANCE_A: 0.35 0.40 0.35
−| Selection |−
GENERATIONS: 15
INDIVIDUALS: 50 0.2 400 0.2
PROGENY: 1
SELECTION: index_ebv high
EBV_METHOD: ssgblup
GENOTYPE_STRATEGY: 10 1.0 parents_offspring 1.0 parents_offspring
INDEX_PROPORTIONS: 0.25 0.75
CULLING: index_ebv 5
-| Mating |-
MATING: random125 simu_anneal
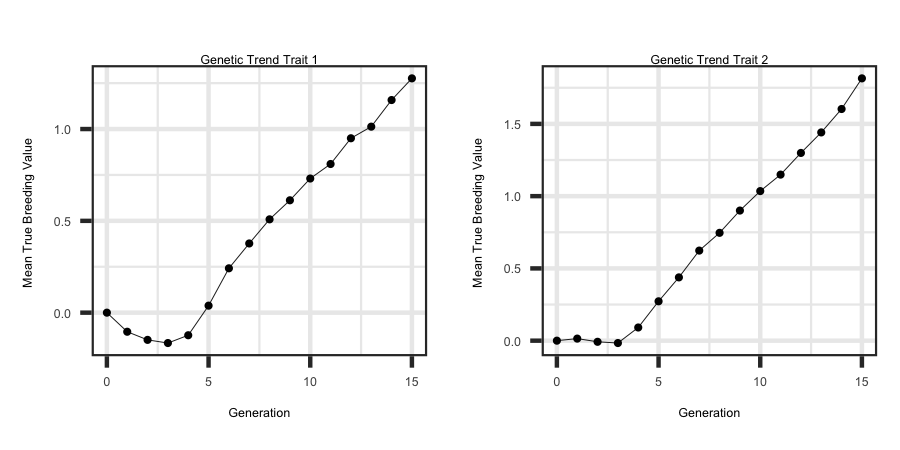
Utilizing the R code outlined below the following plots were generated from the output files to see the response across both traits.
R-Code
rm(list = ls()); gc()
library(ggplot2); library(tidyverse)
setwd("/Users/jeremyhoward/Desktop/C++Code/18_GenoDiver_V3/GenoDiverFiles/")
df <- read_table2(file="Summary_Statistics_DataFrame_Performance",col_names = TRUE,col_type = "dccccccccccccc") %>%
mutate(tbv1 = as.numeric(matrix(unlist(strsplit(tbv1, "[()]")), ncol = 2, byrow = TRUE)[, 1]),
tbv2 = as.numeric(matrix(unlist(strsplit(tbv2, "[()]")), ncol = 2, byrow = TRUE)[, 1])) %>%
select(Generation,tbv1,tbv2)
ggplot(df, aes(x = Generation, y = tbv1)) + geom_line(size = 1) + ggtitle("Genetic Trend Trait 1") + theme_bw() +
theme(plot.title = element_text(hjust = 0.5)) + ylab("Mean True Breeding Value ")
ggplot(df, aes(x = Generation, y = tbv2)) + geom_line(size = 1) + ggtitle("Genetic Trend Trait 2") + theme_bw() +
theme(plot.title = element_text(hjust = 0.5)) + ylab("Mean True Breeding Value ")