The MaCS program (Markovian Coalescence Simulator; Chen et al. 2009) generates the founder genome. Before using the program, it is advisable to understand the coalescent process and a good review paper is Hudson (1991) and Chapter 5 of Charlesworth & Charleworth (2010). We have cho- sen to employ a coalescent simulator (specifically MaCS) in this step due to the flexibility of the approach in generating haplotype sequences for a wide range of population scenarios in terms of the size and structure of the an- cestral population across time and genome scenarios with varying mutation and recombination rates. We have chosen the default scenarios to resemble options specified by AlphaSim (Hickey & Gorjanc, 2012) as they represent typical agricultural species LD patterns.
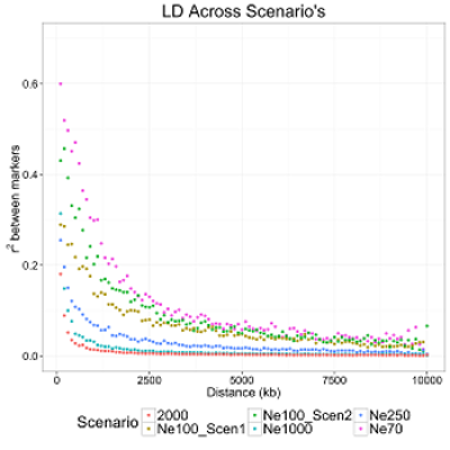
There are 5 default scenarios that represent a range of linkage disequilibrium (LD) patterns that can be called within the simulation, as outlined in the figure below. The five scenarios are called by specifying either 'Ne70', 'Ne100_Scen1', 'Ne100_Scen2', 'Ne250' or 'Ne1000' after the FOUNDER Effective Size parameter in the parameter file. The user can input a custom effective population size and historical population parameters by using 'CustomNe' as the parameter. An easy way to generate custom parameters for MaCS is to utilize a default scenario that resembles the pattern you are wanting and to change the effect population size of the population and determine how the LD pattern changes. If this is specified the program looks for a file called “CustomNe” within the folder where the execution of the program occurred. The file should contain two rows, with the first one being the effective population size parameter and the last one being the historical population size parameters. Lastly, if the program only reads an integer value, then the value is the effective population size with no population history.
The generation of sequence information may take some time to compute. The files generated from the program may be large. Due to this, it is advisable only to generate sequence data once for a given scenario and then adjust narrow-sense heritability, broad sense heritability, selection or mating parameters and start with generating the founder generation.
The default scenarios are below. An illustration of the LD decay associated with each scenario is on the following page:

