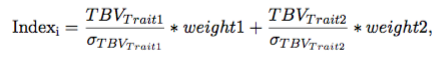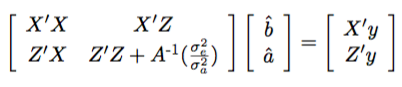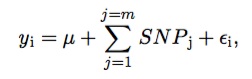Selection and Culling Parameters
GENERATIONS
- Description: Determines the number of generations to simulate and includes the number of generations chosen with the MALE FEMALE FOUNDER option.
- Option: Integer value.
- Usage: '
GENERATIONS: 10'.
- Type: Mandatory.
- Note:
The number of generations allowed vary depending on the size of your computer memory. It is advisable to start at a small number of generations and then build up to determine how much memory a scenario will use.
INDIVIDUALS
- Description: Determines the number of males and females in each generation and replacement rate for parents each generation. If the number of parents falls below the user specified male or female value, the program exits.
- Options:
- Integer value: Number of male parents within each generation.
- Double value (0.0 to 1.0): Proportion of male parents that are culled and replaced by progeny each generation.
- Integer value: Number of female parents within each generation.
- Double value (0.0 to 1.0): Proportion of female parents that are culled and replaced by progeny each generation.
- Usage: '
INDIVIDUALS: 50 0.2 600 0.2'.
- Type: Mandatory.
PARITY_MATES_DIST
- Description: Determines the distribution of the number of mating pairs a sire has for each age group. A Beta (Alpha, Beta) is used to generate the distribution of mating pairs for a given age group. To generate the number of mating pairs by age class, the cumulative distribution function (CDF) is split into quadrants based on the number of age classes that occur within a generation. The total number of mating pairs within an age class is the proportion that falls within the CDF quadrant for a given age class.
- Options:
- Double value (> 0.0): Alpha parameter in Beta distribution.
- Double value (> 0.0): Beta parameter in Beta distribution.
- Usage: '
PARITY_MATES_DIST: 1 1'.
- Type: Optional. The default is both parameters being 1, which is very similar to a uniform distribution, such that all age classes have the same proportion of mating pairs.
PROGENY
- Description: Determines the number of progeny produced by each mating pair. The number of progeny per mating pair may not be the actual number of progeny produced if fitness QTL segregates in the population. The progeny of a mating pair makes it to breeding age if the fitness value of the progeny is greater than a random value derived from a uniform distribution ranging from 0 to 1. The fitness value of the progeny is a function of the multiplicative effect of all lethal and sub-lethal QTL genotype effects.
- Option: Integer value.
- Usage: 'PROGENY: 4'.
- Type: Mandatory.
MAXFULLSIB
- Description: - Determines the maximum number of full-sib progeny selected within a family. Once the maximum number is reached the full-sib with the lowest selection criteria is no longer selected and the next best animal is selected. This process is repeated across all families until all families are below the value.
- Option: Ranges from 1 to number of progeny.
- Usage: 'MAXFULLSIB: 2'.
- Type: Optional. Default set at number of progeny.
SELECTION
- Description: The metric used to select individuals and favorable direction.
- Options:
- Selection Criteria:
- random: random selection.
- phenotype: phenotypic selection.
- tbv: selection based on true breeding value (TBV).
- ebv: selection based on estimated breeding value (EBV).
- index_tbv: selection based on an index for the TBV across both traits.
- index_ebv: selection based on an index for the EBV across both traits.
- Direction:
- high: High values are favorable.
- low: Low values are favorable.
- Usage: '
SELECTION: ebv high'.
- Type: Mandatory.
- Note:
For random selection, the direction does not impact the results and therefore the direction option does not matter. For index tbv and index ebv two traits have to be simulated. The index for animal ‘i’ based on TBV is constructed as follows:
where weight1 and weight2 are the index weights for Trait1 and Trait2, respectively. The index based on EBV is the same except EBV are used instead of TBV. The standard deviation for Trait 1 and 2 is calculated from animals born a generation before truncation selection begins.
INDEX_PROPORTIONS
- Description: Sets the weight that each trait is given when constructing the index true or estimated breeding value when selection is based on index tbv or index ebv.
- Options:
- Double value: Weight given to Trait 1.
- Double value: Weight given to Trait 2.
- Usage: '
INDEX PROPORTIONS: 0.20 0.80'.
- Type: Mandatory if selection based on ‘index tbv’ or ‘index ebv’.
- Note:
The index weights must be such that their sum is equal to 1 and each weight is between 0.0 and 1.0.
EBV_METHOD
- Description: - Parameter that specifies how estimated breeding values (EBV) are generated.
- Options:
- Method:
-
pblup: The best linear unbiased prediction (BLUP) of EBV are obtained by Henderson’s (Henderson 1975) mixed model equations. The mixed model equations are outlined below:
where X and Z refers to fixed and random design matrices, which relate records to fixed and random effects, A
-1 refers to the inverse of the pedigree based relationship matrix. Fixed effects include the intercept and random effects include the effect of the animal. The mixed model equations are solved by the preconditioned conjugate gradient method or cholesky decomposition of the LHS of the matrix.
-
gblup: The same as the pblup option, EBV are obtained by solving the mixed model equations, except the inverse of the pedigree based relationship matrix is replaced by the inverse of the genomic based relationship matrix.
-
rohblup: The same as the pblup option, EBV are obtained by solving the mixed model equations, except the inverse of the pedigree based relationship matrix is re- placed by the inverse of the run-of-homozygosity based relationship matrix.
-
ssgblup: The same as the pblup option, EBV are obtained by solving the mixed model equations, except the inverse of the pedigree based relationship matrix is replaced by the inverse of a relationship matrix that augments pedigree relationships across all individuals based on contributions from animals with genomic information (H
-1; Aguilar et al., 2010; Christensen & Lund, 2010).
-
bayes: Marker effects and the associated EBV are estimated utilizing bayesian whole genome regression models based on the following model:
where y is the phenotype for individuali, μ is the intercept, SNP is the additive genetic effects that correspond to allele substitution effects for each marker and ε is the residual for individual i. SNP covariates had values of 0 for the homozygote, 1 for the heterozygote and 2 for the alternative homozygote. The intercept is assigned an uninformative prior and the SNP covariates can have 4 possi- ble prior densities: 1.) Gaussian (BayesRidgeRegression); 2.) Scaled-t (BayesA); 3.) Two finite mixture priors: a mixture of a point of mass at zero and a Gaussian slab (BayesC); 4.) Two finite mixture priors: a mixture of a point of mass at zero and a Scaled-t slab (BayesB). Sam- ples were drawn from the posterior density using a Gibbs sampler with scalar updating.
- Usage: '
EBV_METHOD: pblup'.
- Type: Only mandatory if selection is based on ebv.
- Note:
If left out and selection is not based on ebv the program will not calculate breeding values. If parameter included and selection is not based on ebv, the program will calculate breeding values.
BLUP_OPTIONS
- Description: - When EBV are estimated based on ’pblup’, gblup’, ’rohblup’ or ’ssgblup’ this parameter can be included to remove older individuals from the analysis, specify solving method and how the inverse of the relationship matrix is calculated.
- Options:
- Generations (integer): Number of generations to trace back from the current group of selection candidates. All animals ’n’ generations back and all of the progeny of the associated parents will be included in the analysis. For example, if a value of 1 is specified the selection candidate’s parents and all progeny that were parents of the selection candidates parents will only be included in the analysis. The default value is the number of generations simulated (i.e. all animals are used each generation).
- Solver:
- direct: Uses Cholesky decomposition for matrix inversion. If EBV accuracy is needed or only simulating a small number of generations this is advised.
- pcg: Uses the iterative preconditioned conjugate gradient (PCG) method and is faster than direct when the number of animals is large. The EBV accuracy is not calculated (Default).
- Genomic Inverse (only applicable if ’gblup’ is specified):
- cholesky: Update previous Cholesky inverse (Meyer et al. 2012; Default).
- recursion: Sequential update (Misztal et al. 2014).
- Usage: '
BLUP_OPTIONS: 4 pcg cholesky'.
- Type: Optional.
- Note:
Parameters that specify inverse calculations are for genomic-based relationships. Calculation of pedigree-based relationships is generated based on Meuwissen & Luo (1992).
G_OPTIONS
- Description: - When ebv are estimated based on 'gblup’ this parameter can be included to determine how the genomic relationship matrix is calculated.
- Options:
- How genomic relationship is constructed:
- VanRaden: Constructed based on G=ZZ’/ 2Σ(pq).
- Allele frequencies calculated:
- founder: Frequencies calculated based on founder genome.
- observed: Frequencies calculated based on animals who are selection candidates.
- Usage: '
G_OPTIONS: VanRaden observed'.
- Type: Optional.
BLENDING_GA22
- Description: - When using the ‘ssgblup’ option, a weighted genomic relationship matrix (Gw), as proposed by VanRaden (2008), is utilized when constructing the H matrix. Gw is constructed as Gw = weight1 * G + weight * A22, where G is the raw genomic relationship matrix and A22 is the pedigree-based relationship for genotyped animals.
- Options:
- Double value: Weight given to the G.
- Double value: Weight given to the A22.
- Usage: '
BLENDING GA22: 0.95 0.05'.
- Type: Optional. Default values are 0.95 for G and 0.05 for A22.
BAYESOPTIONS
- Description: - When ebv are estimated based on the “bayes” option, this parameter has to be included to determine which method to utilize and MCMC options.
- Options:
- Bayesian prediction method: (Required)
- BayesRidgeRegression: Gaussian prior.
- BayesA: Scaled-t prior.
- BayesB: Spike with a point of mass at 0 and slab with the slab having a scaled-t prior.
- BayesC: Spike with a point of mass at 0 and slab with the slab having a gaussian prior.
- Integer value: Number of MCMC iterations the first time a method is implemented. (Required)
- Integer value: Number of MCMC iterations to burn-in the first time a method is implemented. (Required)
- Integer value: Number of MCMC iterations for any generation after the first time a method is implemented. (Required)
- Integer value: Number of MCMC iterations to burn-in for any generation after the first time a method is implemented. (Required)
- Integer value: Thinning rate for MCMC iterations (Required).
- How pie (i.e. proportion of markers with non-null effect) is determined: (Only needed for BayesB & BayesC)
- fix: Fix proportion of SNP with non-null effect.
- estimate: Estimate proportion of SNP with non-null effect.
- Double value: Proportion of markers with non-null effect. (Only needed for BayesB & BayesC)
- Integer value: Number of generations to trace back from the current group of selection candidates. All animals ’n’ generations back and their associated progeny will be included in the analysis. For example, if a value of 1 is specified the selection candidate’s parents and the associated parents progeny will only be included in the analysis. The default value is the number of generations simulated (i.e. all animals are used each generation). (Optional).
- Usage: '
BAYESOPTIONS: BayesC 10000 2000 6000 1000 5 fix 0.10'.
- Type: If 'bayes' EBV option is utilized has to be included otherwise can't be in parameter file.
- Note:
The number of iterations to run can be investigated by looking at the 'bayes_mcmc_samples' files within the output directory. For each generation the files gets
replaced by the most recent results.
CULLING
- Description: The metric used to cull parents and maximum age an animal can remain in the population before being removed due to old age. The direction is the same as the selection direction.
- Options:
- Culling Criteria:
- random: random culling.
- phenotype: phenotypic culling.
- tbv: culling based on true breeding value (TBV).
- ebv: culling based on estimated breeding value (EBV).
- index_tbv: culling based on an index for the TBV across both traits.
- index_ebv: culling based on an index for the EBV across both traits.
- Age Removed:
- Integer: Age at which an animal has to be removed from the population.
- Usage: '
CULLING: ebv 5'.
- Type: Mandatory.
INTERIM_EBV
- Description: Determines whether to calculate interim estimated breeding values (EBV) before or after culling of parents.
- Options:
- no: Interim EBV are not calculated.
- before_culling: Interim EBV calculated prior to culling parents.
- after_culling: Interim EBV calculated after culling parents.
- Usage: '
INTERIM EBV: no'.
- Type: Optional. Default is ‘no’.
PHENOTYPE_STRATEGY
- Description: - When simulating 1 quantitative trait, determines when and if an animal has a phenotype when estimated breeding value (EBV) are being predicted.
- Options:
- Male Proportion Phenotyped:
- Double value (0.0-1.0): Proportion of males that are phenotyped based on the male phenotyping strategy.
- Male Phenotyping Strategy:
- pheno_atselection: Phenotype selection candidates prior to predicting EBV.
- pheno_afterselection: Phenotype selection candidates after predicting EBV.
- pheno_parents: Only phenotype selection candidates that were selected to serve as parents.
- random_atselection: Randomly phenotype selection candidates prior to predicting EBV.
- random_afterselection: Randomly phenotype selection candidates after predicting EBV.
- random_parents: Randomly phenotype selection candidates that were selected to serve as parents.
- ebv_atselection: Phenotype selection candidates based on EBV prior to predicting EBV.
- ebv_afterselection: Phenotype selection candidates based on EBV after predicting EBV.
- ebv_parents: Phenotype selection candidates that were selected based on EBV to serve as parents.
- litterrandom_atselection: Randomly phenotype within each litter prior to predicting EBV.
- litterrandom_atselection: Randomly phenotype within each litter after predicting EBV.
- Female Proportion Phenotyped:
- Double value (0.0-1.0): Proportion of females that are phenotyped based on the female phenotyping strategy.
- Female Phenotyping Strategy:
- pheno_atselection: Phenotype selection candidates prior to predicting EBV.
- pheno_afterselection: Phenotype selection candidates after predicting EBV.
- pheno_parents: Only phenotype selection candidates that were selected to serve as parents.
- random_atselection: Randomly phenotype selection candidates prior to predicting EBV.
- random_afterselection: Randomly phenotype selection candidates after predicting EBV.
- random_parents: Randomly phenotype selection candidates that were selected to serve as parents.
- ebv_atselection: Phenotype selection candidates based on EBV prior to predicting breeding values.
- ebv_afterselection: Phenotype selection candidates based on EBV after predicting breeding values.
- ebv_parents: Phenotype selection candidates that were selected based on EBV to serve as parents.
- litterrandom_atselection: Randomly phenotype within each litter prior to predicting EBV.
- litterrandom_atselection: Randomly phenotype within each litter after predicting EBV.
- Distribution sampling from when doing EBV based phenotype strategy:
- high: Phenotype animals with a high EBV.
- low: Phenotype animals with a low EBV.
- tails: Phenotype animals with a high and low EBV.
- Usage: '
PHENOTYPE_STRATEGY: 1.0 atselection 1.0 atselection'.
- Type: Optional. The default setting is all male and female selection candidates are phenotyped prior to selection.
- Note:
For the pheno_* the proportion has to be 0.0 or 1.0, while any other scenario has to be greater than 0.0 and less than 1.0. Both male and female proportions can’t be 0.0.
PHENOTYPE_STRATEGY1
- Description: - When simulating 2 quantitative traits, determines when and if an animal has a phenotype when estimated breeding value (EBV) are being predicted for trait 1.
- Options:
- Male Proportion Phenotyped:
- Double value (0.0-1.0): Proportion of males that are phenotyped based on the male phenotyping strategy.
- Male Phenotyping Strategy:
- pheno_atselection: Phenotype selection candidates prior to predicting EBV.
- pheno_afterselection: Phenotype selection candidates after predicting EBV.
- pheno_parents: Only phenotype selection candidates that were selected to serve as parents.
- random_atselection: Randomly phenotype selection candidates prior to predicting EBV.
- random_afterselection: Randomly phenotype selection candidates after predicting EBV.
- random_parents: Randomly phenotype selection candidates that were selected to serve as parents.
- ebv_atselection: Phenotype selection candidates based on EBV prior to predicting EBV.
- ebv_afterselection: Phenotype selection candidates based on EBV after predicting EBV.
- ebv_parents: Phenotype selection candidates that were selected based on EBV to serve as parents.
- litterrandom_atselection: Randomly phenotype within each litter prior to predicting EBV.
- litterrandom_atselection: Randomly phenotype within each litter after predicting EBV.
- Female Proportion Phenotyped:
- Double value (0.0-1.0): Proportion of females that are phenotyped based on the female phenotyping strategy.
- Female Phenotyping Strategy:
- pheno_atselection: Phenotype selection candidates prior to predicting EBV.
- pheno_afterselection: Phenotype selection candidates after predicting EBV.
- pheno_parents: Only phenotype selection candidates that were selected to serve as parents.
- random_atselection: Randomly phenotype selection candidates prior to predicting EBV.
- random_afterselection: Randomly phenotype selection candidates after predicting EBV.
- random_parents: Randomly phenotype selection candidates that were selected to serve as parents.
- ebv_atselection: Phenotype selection candidates based on EBV prior to predicting breeding values.
- ebv_afterselection: Phenotype selection candidates based on EBV after predicting breeding values.
- ebv_parents: Phenotype selection candidates that were selected based on EBV to serve as parents.
- litterrandom_atselection: Randomly phenotype within each litter prior to predicting EBV.
- litterrandom_atselection: Randomly phenotype within each litter after predicting EBV.
- Distribution sampling from when doing EBV based phenotype strategy:
- high: Phenotype animals with a high EBV.
- low: Phenotype animals with a low EBV.
- tails: Phenotype animals with a high and low EBV.
- Usage: '
PHENOTYPE_STRATEGY1: 1.0 atselection 1.0 atselection'.
- Type: Optional. The default setting is all male and female selection candidates are phenotyped prior to predicting breeding values for the first trait.
- Note:
For the pheno_* the proportion has to be 0.0 or 1.0, while any other scenario has to be greater than 0.0 and less than 1.0. Both male and female proportions can’t be 0.0.
PHENOTYPE_STRATEGY2
- Description: - When simulating 2 quantitative traits, determines when and if an animal has a phenotype when estimated breeding value (EBV) are being predicted for trait 2.
- Options:
- Male Proportion Phenotyped:
- Double value (0.0-1.0): Proportion of males that are phenotyped based on the male phenotyping strategy.
- Male Phenotyping Strategy:
- pheno_atselection: Phenotype selection candidates prior to predicting EBV.
- pheno_afterselection: Phenotype selection candidates after predicting EBV.
- pheno_parents: Only phenotype selection candidates that were selected to serve as parents.
- random_atselection: Randomly phenotype selection candidates prior to predicting EBV.
- random_afterselection: Randomly phenotype selection candidates after predicting EBV.
- random_parents: Randomly phenotype selection candidates that were selected to serve as parents.
- ebv_atselection: Phenotype selection candidates based on EBV prior to predicting EBV.
- ebv_afterselection: Phenotype selection candidates based on EBV after predicting EBV.
- ebv_parents: Phenotype selection candidates that were selected based on EBV to serve as parents.
- litterrandom_atselection: Randomly phenotype within each litter prior to predicting EBV.
- litterrandom_atselection: Randomly phenotype within each litter after predicting EBV.
- Female Proportion Phenotyped:
- Double value (0.0-1.0): Proportion of females that are phenotyped based on the female phenotyping strategy.
- Female Phenotyping Strategy:
- pheno_atselection: Phenotype selection candidates prior to predicting EBV.
- pheno_afterselection: Phenotype selection candidates after predicting EBV.
- pheno_parents: Only phenotype selection candidates that were selected to serve as parents.
- random_atselection: Randomly phenotype selection candidates prior to predicting EBV.
- random_afterselection: Randomly phenotype selection candidates after predicting EBV.
- random_parents: Randomly phenotype selection candidates that were selected to serve as parents.
- ebv_atselection: Phenotype selection candidates based on EBV prior to predicting breeding values.
- ebv_afterselection: Phenotype selection candidates based on EBV after predicting breeding values.
- ebv_parents: Phenotype selection candidates that were selected based on EBV to serve as parents.
- litterrandom_atselection: Randomly phenotype within each litter prior to predicting EBV.
- litterrandom_atselection: Randomly phenotype within each litter after predicting EBV.
- Distribution sampling from when doing EBV based phenotype strategy:
- high: Phenotype animals with a high EBV.
- low: Phenotype animals with a low EBV.
- tails: Phenotype animals with a high and low EBV.
- Usage: '
PHENOTYPE_STRATEGY2: 1.0 atselection 1.0 atselection'.
- Type: Optional. The default setting is all male and female selection candidates are phenotyped prior to predicting breeding values for the second trait.
- Note:
For the pheno_* the proportion has to be 0.0 or 1.0, while any other scenario has to be greater than 0.0 and less than 1.0. Both male and female proportions can’t be 0.0.
GENOTYPE_STRATEGY
- Description: - If doing ‘ssgblup’ based BLUP determines who and at what time an animal is genotyped.
- Options:
- Generation (integer value): Generation to begin genotyping.
- Male Proportion Genotyped:
- Double value (0.0-1.0): Proportion of males that are genotyped based on the male genotyping strategy.
- Male Genotyping Strategy:
- parents: Genotype all parents.
- offspring: Genotype all selection candidates.
- parents_offspring: Genotype all parents and selection candidates.
- random: Randomly genotype a proportion of the selection candidates.
- parents_random: Randomly genotype a proportion of the selection candidates and all selection candidates that were selected to become parents.
- ebv: Genotype a proportion of the selection candidates based on EBV.
- parents_ebv: Genotype a proportion of the selection candidates based on EBV and all selection candidates that were selected to become parents.
- litter_random: Genotype a random proportion of the animals within each litter.
- litter_parents_random: Genotype a random proportion of the animals within each litter and all selection candidates that were selected to become parents.
- Female Proportion Genotyped:
- Double value (0.0-1.0): Proportion of females that are genotyped based on the female genotyping strategy.
- Female Genotyping Strategy:
- parents: Genotype all parents.
- offspring: Genotype all selection candidates.
- parents_offspring: Genotype all parents and selection candidates.
- random: Randomly genotype a proportion of the selection candidates.
- parents_random: Randomly genotype a proportion of the selection candidates and all selection candidates that were selected to become parents.
- ebv: Genotype a proportion of the selection candidates based on EBV.
- parents_ebv: Genotype a proportion of the selection candidates based on EBV and all selection candidates that were selected to become parents.
- litter_random: Genotype a random proportion of the animals within each litter.
- litter_parents_random: Genotype a random proportion of the animals within each litter and all selection candidates that were selected to become parents.
- Distribution sampling from when doing EBV based genotyping strategy:
- high: Genotype animals with a high EBV.
- low: Genotype animals with a low EBV.
- tails: Genotype animals with a high and low EBV.
- Usage: '
GENOTYPE_STRATEGY: 1.0 parents 1.0 parents'.
- Type: Mandatory if doing ‘ssgblup’ option.
- Note:
For the parents, offspring and parents_offspring option the proportion has to be 0.0 or 1.0, while any other scenario has to be greater than 0.0 and less than 1.0. The distribution to sample is not required and if not provided it will be the direction animals are selected on.